CRISPR-Sirius, a sensitive and bright imaging system, has been developed by research groups led by Dr. Ma Hanhui at ShanghaiTech University and Dr. Thoru Pederson at the University of Massachusetts Medical School. Their study was recently published in Nature Methods, a prestige scientific journal.
CRISPR-Sirius makes it possible to detect low-copy repeats distributed in the intergenic and genic regions and to track the dynamics of multiple genomic elements from kilobases to megabases apart. Dr. Ma has engineered CRISPR guide RNA scaffolds for signal amplification in genome imaging, collaborating with Dr. Zhang Shaojie’s group at the University of Central Florida.
The human genome contains 3 billion base pairs with an approximately 2-meter length in the majority of cells in a human body. It is still unclear how the chromatin DNAs are packed into a nucleus with a diameter of 10 to 20 micrometers and orchestrated by dynamic interactions in living cells. CRISPR has been repurposed for imaging chromatin dynamics in live cells. However, the previously-developed methods rely heavily on the rare high-copy repeats with limit biological applications in the human genome.
Previously, Dr. Ma has developed CRISPR-Broccoli to monitor guide RNA stability in living cells. Here the CRISPR-Broccoli technology reveals that guide RNAs produced by inserting RNA aptamers into their tetraloop are much more stable than those by tagging RNA aptamers to their 3'-terminus (Figure 1a, 1b).
Now, Dr. Ma and collaborators have developed the CRISPR-Sirius technology by designing stable octet of RNA aptamers rationally and then inserting them into the tetraloop of the guide RNA scaffold (Figure 1c, 1d). This modification increases the brightness and sensitivity significantly for the DNA imaging without disrupting the stability of guide RNAs. Therefore, the enhanced CRISPR-Sirius imaging system provides a platform for a variety of biological applications, including genomic interactions from kilobases to megabases, chromatin organization and dynamics of sister chromatid separation.
Key co-authors of this paper include Tu Li-Chun and David Grunwald from University of Massachusetts Medical School, Ardalan Naseri from University of Central Florida and Chung Yu-Chieh from University of Tokyo. Their work was supported, in part, by ShanghaiTech University and NIH 4D Nucleome Program.
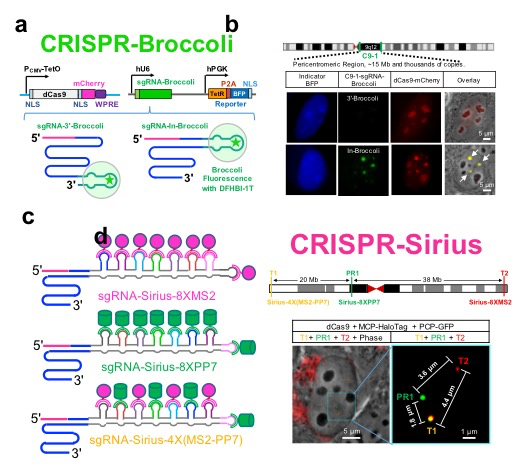
Figure. Development and applications of CRISPR-Sirius. a. Diagram of the CRISPR-Broccoli system; b. Monitoring the stability of CRISPR guide RNA in living cells by CRISPR-Broccoli; c. Diagram of the CRISPR-Sirius system; d. Tracking chromatin dynamics in live cells by CRISPR-Sirius.


