Recently, different research teams led by Professor Sun Bo from SLST and Professor Xi Xu-Guang from French National Centre for Scientific Research (CNRS) have collaboratively published a research article entitled “Human RPA activates BLM's bidirectional DNA unwinding from a nick” in the journal eLife, revealing a novel unwinding model of a DNA-repair-involved helicase.
DNA helicases are a class of enzymes vital to all organisms. They are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two annealed nucleic acid strands using energy from NTP hydrolysis. Among them, the RecQ family helicases are a kind of proteins that are very conserved in the evolutionary process. They play important roles in maintaining the stability of the genome by participating in many cellular processes, such as DNA replication, recombination, and repair. Bloom syndrome helicase (BLM) is one of five mammalian RecQ helicases and functions in many aspects of maintaining genetic stability, including unwinding of different DNA structures, processing of double-stranded DNA break (DSB) ends, and restarting of replication fork.
BLM helicase works with multiple proteins, it processes distinct DNA substrates, but not nicked DNA, during many steps in DNA replication and repair. However, how BLM prepares itself for diverse functions remains elusive. In this study, using a combined single-molecule approach (Figure 1A), the researchers found that a high abundance of BLMs can indeed unidirectionally unwind double-stranded DNA (dsDNA) from a nick when an external destabilizing force is applied. Strikingly, human replication protein A (hRPA) not only ensures that limited quantities of BLMs unwind nicked dsDNA under a reduced force but also permits the translocation of BLMs on both intact and nicked single-stranded DNA (ssDNA), resulting in a bidirectional unwinding mode (Figure 1B). This finding presents novel DNA unwinding activities of BLM that potentially facilitate its function switching in DNA repair. The study also provides insights into the mechanisms by which BLM and RPA cooperate to facilitate end resection in HR. It is highly likely that the abilities of BLM to recognize and preload on the nick would help identify the locations of DSB and possibly play a role in recruiting other required proteins for the resection. In addition, the inherent ability of BLM and RPA to unwind bidirectionally from a nick would facilitate the resection by DNA adduct coated DSB ends which provides ssDNA for nucleases to degrade.
This work was supported by the National Key R&D Program of China, Natural Science Foundation of Shanghai, ShanghaiTech University Startup funding, the French National Cancer Institute, and the French National League Against Cancer.
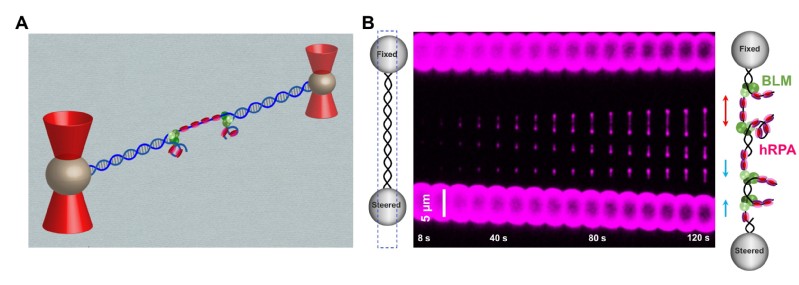
Figure 1: A, A schematic of the experimental configuration; B, Representative kymographs of unidirectional and bidirectional DNA unwinding.
Read more at:https://elifesciences.org/articles/54098


