In a News and Views commentary entitled To BE or not to BE, that is the question, Dr. Jia Chen at School of Life Science and Technology (SLST) of ShanghaiTech University, together with Dr. Bei Yang at Shanghai Institute for Advanced Immunochemical Studies (SIAIS) of ShanghaiTech University and Dr. Li Yang at CAS-MPG Partner Institute for Computational Biology (PICB) of Chinese Academy of Sciences, commented the recent studies published in Science and Nature Biotechnology about precision of base editing. The News and Views commentary was published online in Nature Biotechnology on April 18, 2019.
Base editors (BEs) were recently developed by combining different nucleoside deaminase family members with the CRISPR-Cas9 system and have been used for targeted C-to-T/A-to-G base editing in various cells or living organisms. Since BEs catalyze the deamination of cytidine or adenosine at target sites without generating DNA double strand breaks and that the majority of human pathogenic variants are single mutations, BEs were thought to be promising tools to correct these disease-related point mutations. However, recent studies showed that cytosine base editors (especially a commonly-used cytosine base editor, BE3), but not adenine base editors, induce elevated levels of genome-wide off-target substitutions (Zuo et al., 2019, Science; Jin et al., 2019, Science; Kim et al., 2019, Nature Biotechnology), therefore raising concerns about the safety of cytosine base editing.
In this News and Views commentary, the authors indicated that the detected off-target editing may be caused by the rat APOBEC1 moiety of BE3 directly. The authors further hypothesized that the use of native or engineered APOBEC deaminases with relatively low DNA binding or catalytic activity may help to reduce unexpected off-target substitutions. Meanwhile, deamination activity-reduced BEs might lead to inefficient on-target editing. However, high on-target activity is essential for broad applications of base editing, especially for therapeutic-related applications in somatic cells. In this scenario, it will be more prudent to adopt strategies other than using activity-reduced deaminases when developing high-precision BEs going forward.
Paper Link:https://www.nature.com/articles/s41587-019-0119-x
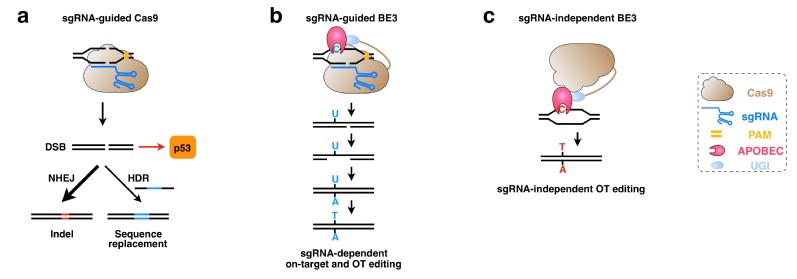
a. Gene editing and p53-mediated DNA damage response induced by Cas9
b. sgRNA-dependent on-target and off-target base editing induced by BE3
c. sgRNA-independent off-target editing induced by BE3


