|
Single Molecule Enzymology Group
Principal investigatorName: Bo SunAssociate Professor , PhD, Associate Professor
Position: Affiliation: School of Life Science and Technology
Honor: Education Background:
Working Experience:
Group Introduction Research Area:
Single Molecule Enzymology, Single Molecule Biophysics
Research Interests:
Ensemble studies have contributed tremendously to comprehending biological reactions. However, these studies characterize the average molecular population, and have limited ability in detecting intermediate states or distinguishing heterogeneities. Over the past few decades, single-molecule techniques, including fluorescence resonance energy transfer (FRET), magnetic tweezers (MT) and optical tweezers (OT), have proven to be exceedingly powerful in addressing this knowledge gap. By studying one molecule at a time, these approaches have enabled significant advancement in the understanding of a wide variety of biomolecular systems, especially those involving DNA and associated proteins. Using these single‐molecule techniques, our research interests are focused on understanding the mechanisms of molecular motors in the process of DNA replication, repair, transcription, and editting, such as helicase, polymerase and nuclease. Group Website:
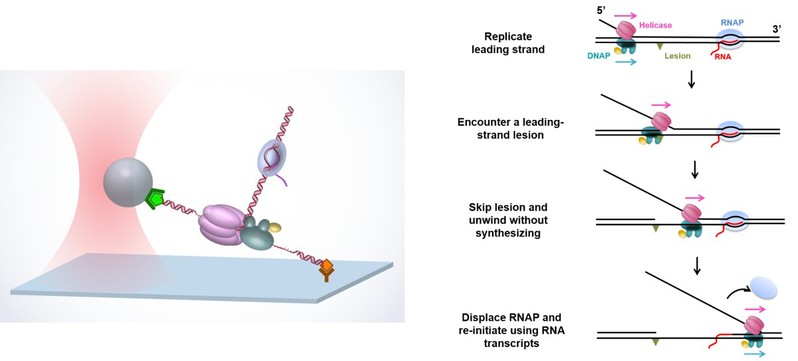
Research Achievement1. An international research collaboration has shed new light on DNA replication. The collaboration, led by Professor Bo Sun from the School of Life Science and Technology at ShanghaiTech University and Professor Michelle D. Wang from the Howard Hughes Medical Institute (HHMI) and Cornell University, revealed a new replication recovery pathway. Their paper, “Helicase Promotes Replication Re-initiation from an RNA transcript,” was published online in Nature Communications on June 13, 2018. This work reveals a novel pathway of replication re-initiation enabled by the participation of a replicative helicase.
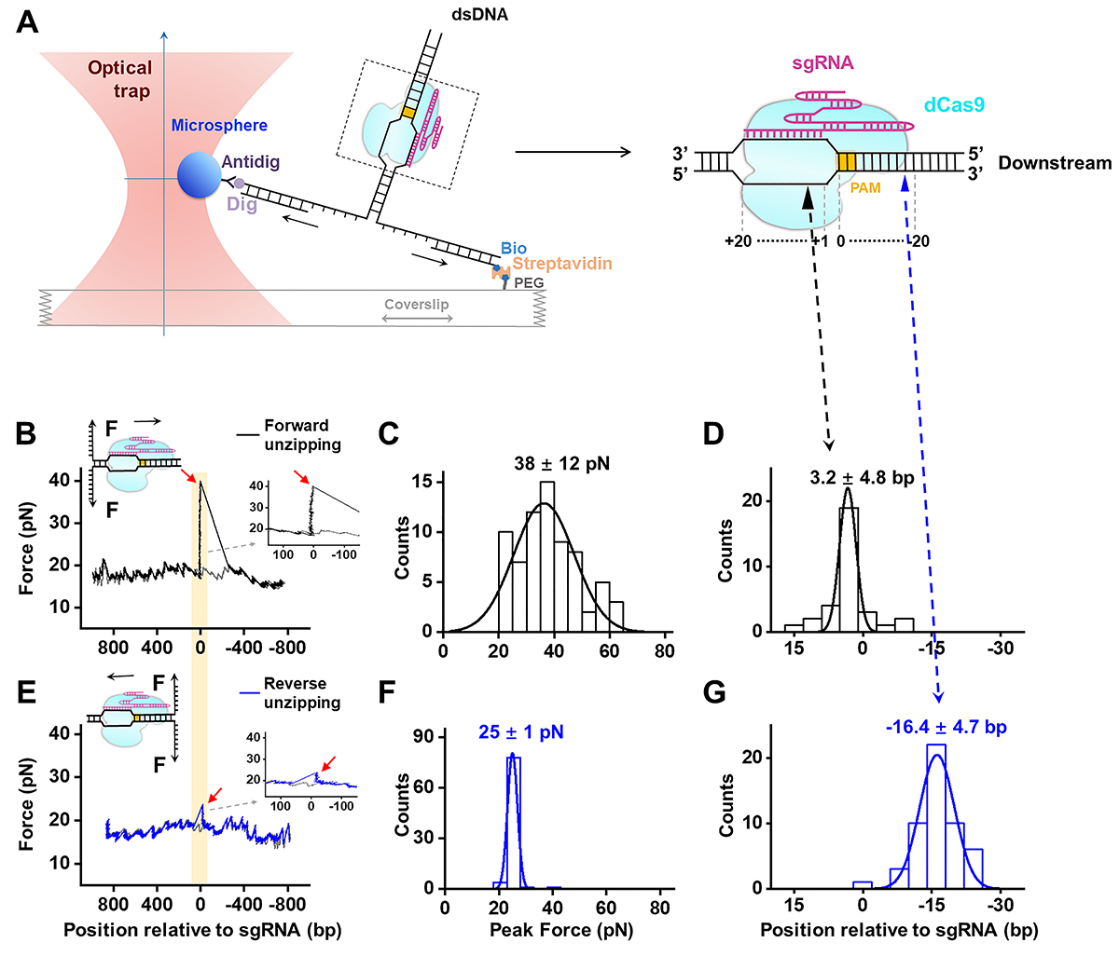
Figure 1. Helicase assists a non-replicating DNAP in bypassing a lesion via RNAP displacement. 2. Research teams led by Professor Sun Bo from SLST and Professor Shen Bin from Nanjing Medical University have completed a high-resolution quantitative map of Cas9/sgRNA/DNA interactions. On November 13, 2019, this work was published as a research article entitled, “The post-PAM interaction of RNA-guided spCas9 with DNA dictates its target binding and dissociation, in the journal Science Advances. This work identifies a critical interaction of Cas9 with DNA that dictates its binding and dissociation, suggesting a distinct strategy to modulate Cas9 activity.
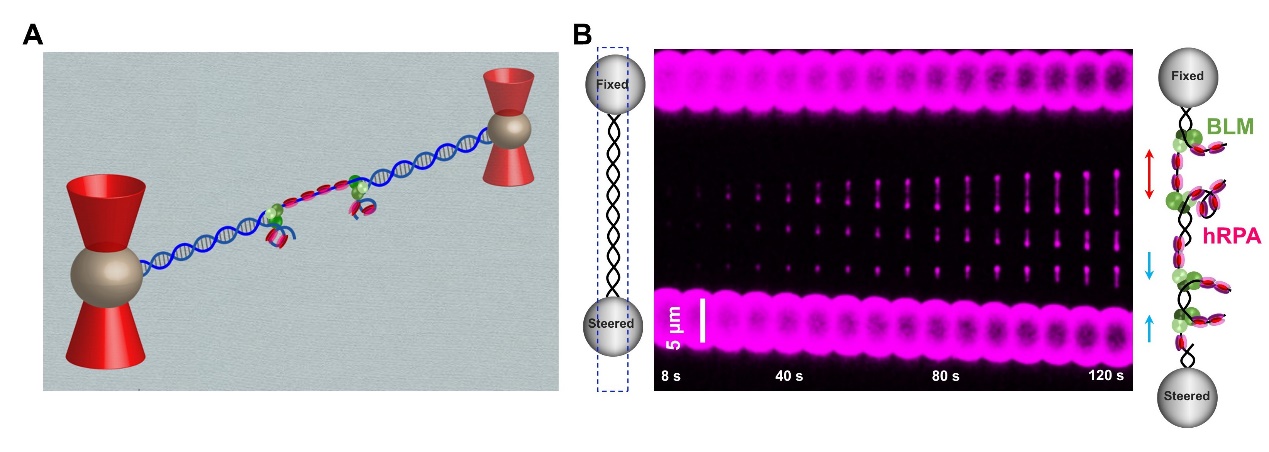
Figure 2. Experimental configuration for single-molecule unzipping experiments and spCas9-sgRNA-DNA interaction mapping. 3. On Feburary 25, 2020, research teams led by Professor Sun Bo from SLST and Professor Xi Xu-Guang from French National Centre for Scientific Research (CNRS) have collaboratively published a research article entitled “Human RPA activates BLM's bidirectional DNA unwinding from a nick” in the journal eLife, revealing a novel unwinding model of a DNA-repair-involved helicase.
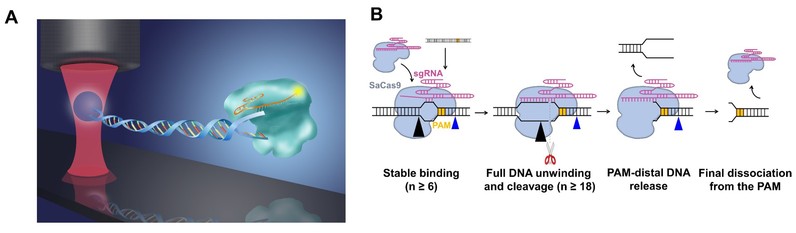
Figure 3. A, A schematic of the experimental configuration; B, Representative kymographs of unidirectional and bidirectional DNA unwinding. 4. Research team led by Professor Bo Sun has systemically investigated the molecular mechanism governing DNA binding, unwinding and cleavage by SaCas9. They provide a detailed dynamic understanding of SaCas9 in DNA target association and dissociation. In addition, two stable interactions between SaCas9 and DNA governing their interplay have been identified. On August 13, 2020, this study was published as a research article entitled, “Dynamics of Staphylococcus aureus Cas9 in DNA target Association and Dissociation”, in the journal EMBO Reports.
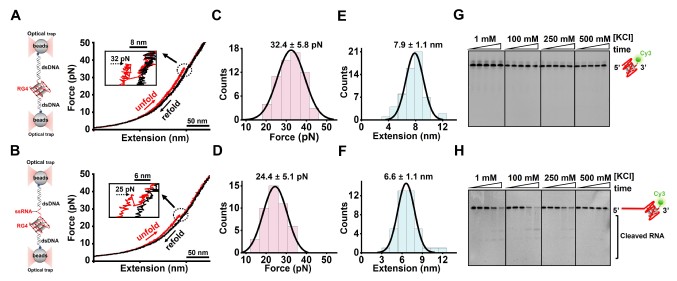
Figure 4. A, A combination of optical tweezer and confocal microscopy to detect the dissociation of SaCas9 after DNA cleavage; B, A proposed model for SaCas9. 5. On March 30, 2021, Sun lab published a research paper entitled "Proximal single-stranded RNA destabilizes human telomerase RNA G-quadruplex and induces its distinct conformers" in The Journal of Physical Chemistry Letters. Using circular dichroism, nuclear magnetic resonance, single-molecule fluorescence resonance energy transfer, single-molecule optical tweezers and enzymatic digestion assays, this work revealed that the stability and conformation of an RNA G-quadruplex (RG4) forming sequence identified near the end of human telomerase RNA is significantly impaired by its proximal ssRNA. This work provides new insights into the stability and folding dynamics of RG4 that are essential for understanding its biological functions.
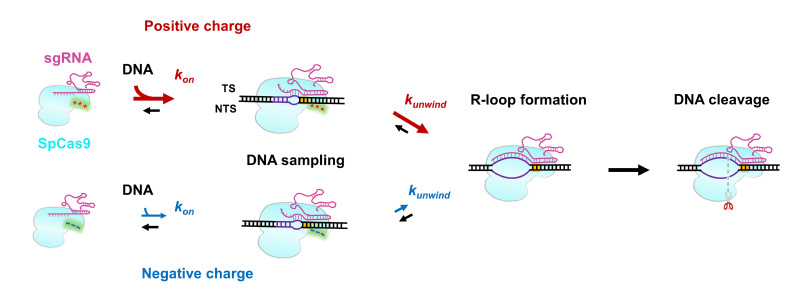
Figure 5. A-H, Mechanical stability and accessibility of the RG4-forming sequence detected by optical tweezers and in vitro enzymatic cleavage assays. 6. On November 25, 2021, Sun lab published a research paper entitled "Efficient DNA interrogation of SpCas9 governed by its electrostatic interaction with DNA beyond the PAM and protospacer" in Nucleic Acids Research. This work identified the nature of an indispendsable interaction site between SpCas9 and DNA. Moreover, the molecular mechanisms and functions of this site in regulating SpCas9 activity have been deciphered.
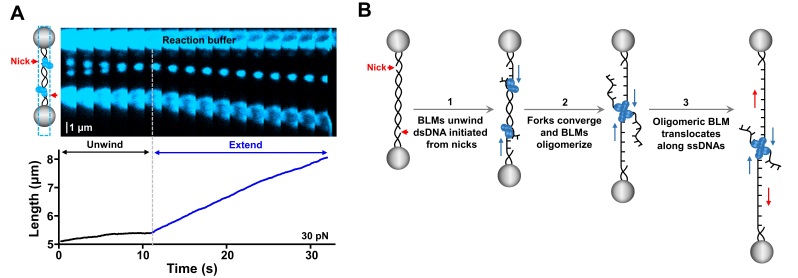
Figure 6. A proposed model for how the post-PAM interaction regulates SpCas9 activity. 7. On June 3, 2022, Sun lab published a research paper entitled, "The convergence of head-on DNA unwinding forks induces helicase oligomerization and activity transition", in PNAS. This work uncovers a novel activity transition pathway for helicases and helps to explain how BLM plays both pro- and anti-recombination roles in the maintenance of genome stability.
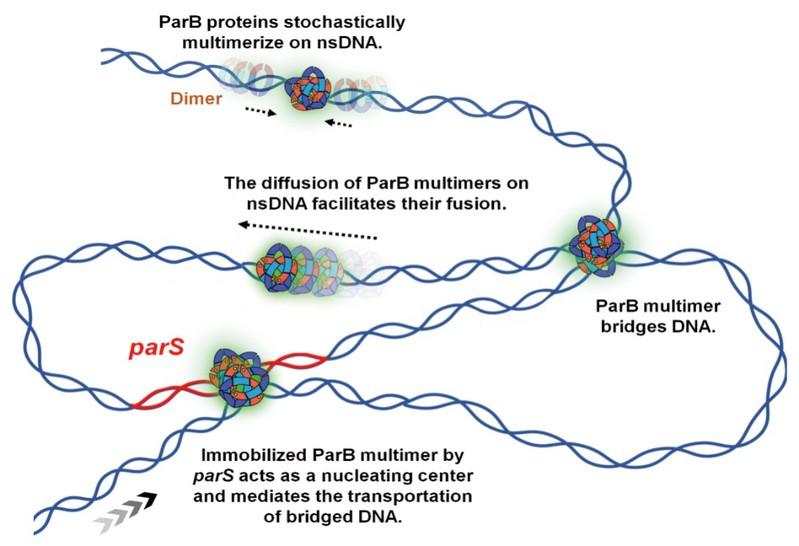
Figure 7. BLM oligomerization leads to an immediate helicase activity transition from dsDNA unwinding to ssDNA translocation. 8. On July 29, 2022, Sun lab published a research paper entitled, "Stochastically multimerized ParB orchastrates DNA assembly as unveiled by single-molecule analysis", in Nucleic Acids Research. This work uncovers unique features of ParB bridging and transporting DNA molecules via multimerization, providing the molecular basis for understanding how the ParABS system mediates chromosome segregation.
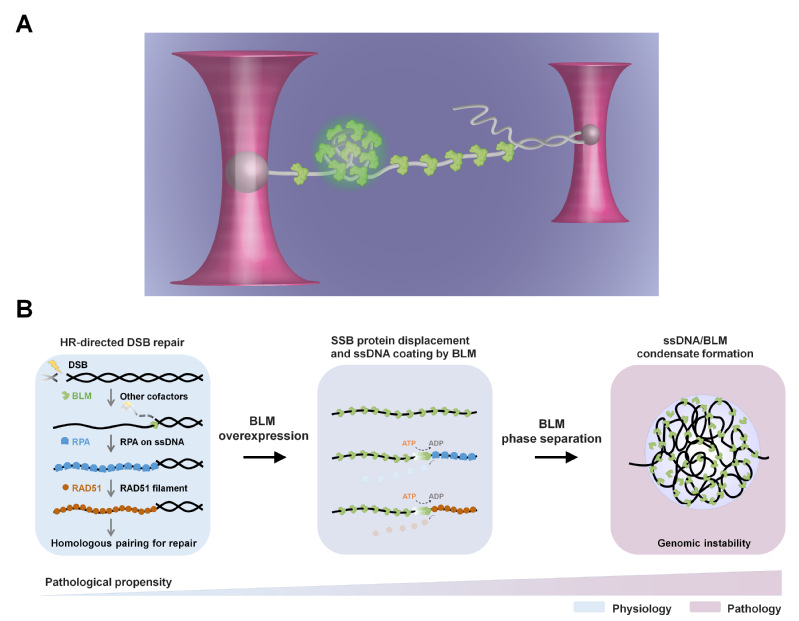
Figure 8. Model of ParB multimer organizating DNA assembly. 9. On August 3, 2022, Sun lab published a research paper entitled, "Bloom syndrome helicase compresses single-stranded DNA into phase-separated condensates", in Angewandte Chemie International Edition. This work reveals that BLM helicase can undergo phase separation and form condensates on ssDNA, giving rise to the compresion and condensation of ssDNA into the condensates.
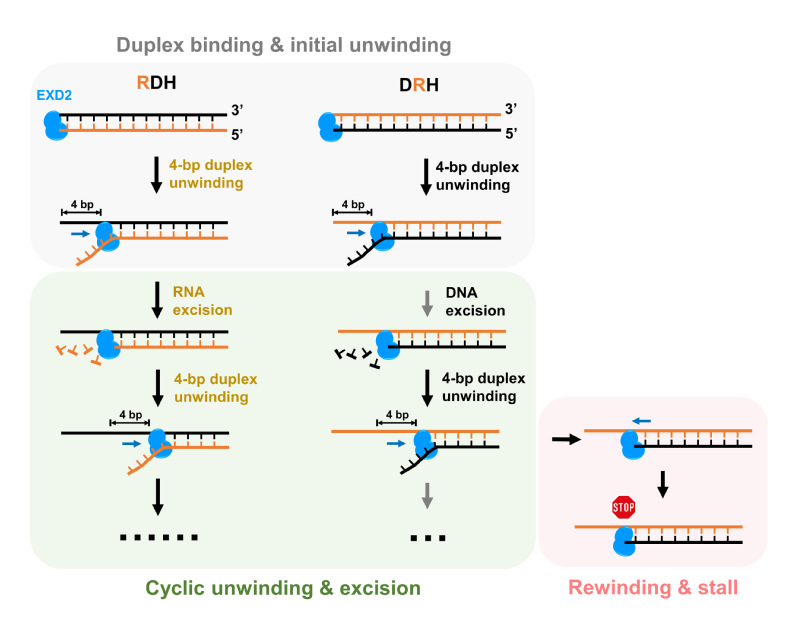
Figure 9. Single-molecule assay and a proposed model of BLM interfering with homologous recombination via ssDNA condensation. 10. On November 3, 2022, Sun lab published a research paper entitled, Discrete RNA–DNA hybrid cleavage by the EXD2 exonuclease pinpoints two rate-limiting steps, in The EMBO Journal. This work reveals that EXD2-mediated hybrid cleavage proceeds in a discrete stepwise pattern, wherein a sudden 4-bp duplex unwinding increment and the subsequent dwell constitute a complete hydrolysis cycle.
Figure 10. A mechanistic model of EXD2 cleaving RNA–DNA hybrids. Representative Publications (*First Author, # Corresponding Author)
MonographBi, Lulu; Qin, Zhenheng; Hou, Xi-Miao; Modesti, Mauro; Sun, Bo. Simultaneous Mechanical and Fluorescence Detection of Helicase-Catalyzed DNA unwinding (Book Chapter). OPTICAL TWEEZERS: METHODS AND PROTOCOLS. Springer, 2022. PatentFunding
AwardsResearch AchievementGroup Member and Photo
|