Systems Biology of Bacterial Drug Tolerance Group
Principal investigator
Name:
Tianmin WangAssistant Professor , PhD, Assistant Professor
Position:
Affiliation:
School of Life Science and Technology
Honor:
Education Background:
- 2008/09-2012/07, Peking University, Bachelor
- 2013/09-2015/07, Tokyo Institute of Technology, Master (THU-TIT exchange program)
- 2012/08-2018/07, Tsinghua University, Ph.D.
Working Experience:
- 2019/01-2023/03, Tsinghua University, School of Medicine, Postdoctoral Researcher
- 2023/05-Now, ShanghaiTech University, School of Life Science and Technology, Assistant Professor (Tenure-Track)
Group Introduction
Research Area:
Bacterial Drug Tolerance, Systems Biology, Spatiotemporal Multiomics, Bacterial Genomics, Biofilm, Microfluidics
Research Interests:
Antibiotics have been one of the most significant medical discoveries in the 20th century and form a crucial foundation for the functioning of modern healthcare systems. However, the current pace of discovering new antibiotics has slowed down, while pathogenic microorganisms are increasingly demonstrating enhanced resistance at both phenotypic and genotypic levels. This has led to the looming antibiotic crisis, which poses a major challenge to public health in this century. In this context, comprehending the molecular mechanisms underlying the generation, development, and evolution of bacterial resistance at different scales can maximize the killing efficacy of existing antibiotics and minimize the probability of resistance emergence. Furthermore, it can aid in the discovery of novel molecular targets to facilitate the design of new drugs, thereby providing a solid scientific foundation for addressing the antibiotic crisis.
Our research group is dedicated to systematically studying the molecular mechanisms of bacterial phenotypic tolerance and the dynamics of genetic resistance evolution at various scales, including the molecular, cellular, and community levels. The laboratory integrates cutting-edge experimental techniques (spatial transcriptomics, high-throughput genetic screening, microfluidics, single-cell technologies, time-lapse fluorescence microscopy, etc) and computational approaches (bacterial comparative genomics, machine learning, stochastic dynamical modeling, etc), aiming to understand the drug tolerance behaviors of diverse pathogens at different scales, with a particular emphasis on unveiling how tolerance (or susceptibility) emerges from the coordination (or dysregulation) of biological networks from a systems biology perspective.During this process, we will also utilize microfluidics, next-generation sequencing, and nanopore sequencing platforms to develop innovative technologies, aiming to provide tools for basic microbiology research, mining novel antimicrobials and enabling rapid diagnosis of drug-resistant bacteria in clinical settings.
Research Achievement
We developed RAINBOW-seq to profile the spatial transcriptome of bacterial community with a resolution of 50 microns, uncovered rich coordination of metabolism within E. coli biofilm (Nat. Chem. Biol. 2023); 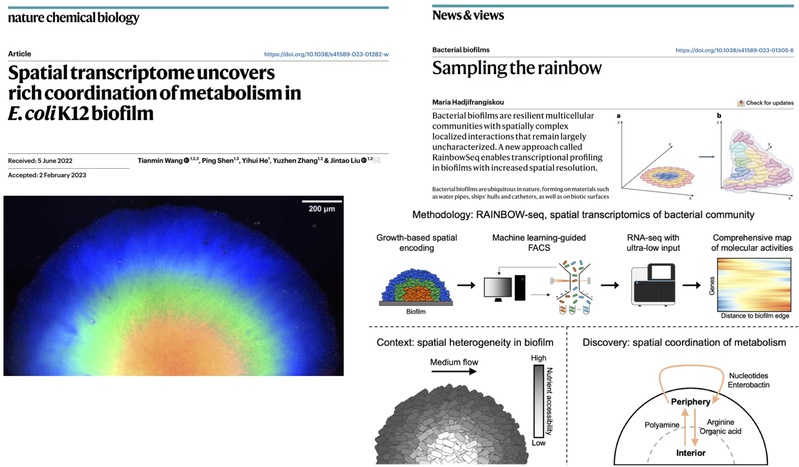
Using spatial transcriptomics, we discovered that, akin to spatially structured mammalian tissues, bacterial community actively maintains spatial homeostasis via latent regulations; and such preserved architecture is crucial for emergent drug tolerance in pathogenic biofilms (in preparation);
Use deep mutational scanning and stochastic modeling, we showed that how precisely coordinated dynamics of transcription-translation modularly tune bacterial indole signaling via TnaC sensor (Nat. Chem. Biol. 2020); 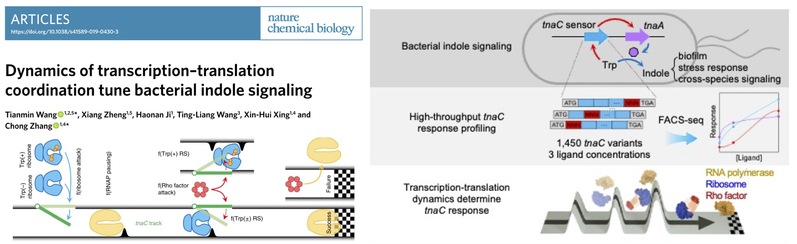
For the first time in bacteria, we established CRISPRi-seq method for functional genomics study in a pooled screen manner, which led to new biology such as tRNA essentiality and stress-tolerance-related gene atlas (Nat. Commun. 2018);
Combining high-throughput screen and machine learning, we decoded the relation of sgRNA sequence with on/off-target activity in CRISPR/Cas9 genome engineering in living bacterial cells, giving rise to predictive models to design active and specific sgRNA library in bacteria (Nucleic Acids Res. 2018, 2021).
Representative Publications (*First Author, # Corresponding Author)
- 1. Zheng, Yayun*; Chai, Ruochen*; Wang, Tianmin#*; Xu, Zeqi; He, Yihui; Shen, Ping; Liu, Jintao#.RNA polymerase stalling-derived genome instability underlies ribosomal antibiotic efficacy and resistance evolution.Nature Communications. 03 August 2024.
- 2. Feng, Huibao#*; Li, Fan; Wang, Tianmin; Xing, Xin-Hui; Zeng, An-Ping; Zhang, Chong; .Deep-learning–assisted Sort-Seq enables high-throughput profiling of gene expression characteristics with high precision.SCIENCE ADVANCES. Nov 2023. 9(45).
- 3. Wang, Tianmin*; Shen, Ping*; He, Yihui; Zhang, Yuzhen; Liu, Jintao#. Spatial transcriptome uncovers rich coordination of metabolism in E. coli K12 biofilm. Nature Chemical Biology. April 2023. 19(8):940-950.
- 4. Wang, Tianmin#*; Shen, Ping*; Chai, Ruochen; He, Yihui; Liu, Jintao#. Profiling of bacterial transcriptome from ultra-low input with MiniBac-seq. Environmental Microbiology. August 2022. 24(12):5774-5787.
- 5. Wang, Tianmin#*; Zheng, Xiang*; Ji, Haonan; Wang, Ting-Liang; Xing, Xin-Hui; Zhang, Chong#. Dynamics of transcription–translation coordination tune bacterial indole signaling. Nature Chemical Biology. April 2020. 16(4):440-449.
- 6. Feng, Huibao*; Guo, Jiahui*; Wang, Tianmin#*; Zhang, Chong#; Xing, Xin-Hui. Guide-target mismatch effects on dCas9–sgRNA binding activity in living bacterial cells. Nucleic Acids Research. February 2021. 49(3):1263-1277.
- 7. Wang, Tianmin*; Guan, Changge*; Guo, Jiahui; Liu, Bing; Wu, Yinan; Xie, Zhen; Zhang, Chong#; Xing, Xin-Hui. Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance. Nature Communications. June 2018. 9(1):2475.
- 8. Wang, Tianmin*; Guo, Jiahui*; Liu, Yangyang*; Xue, Zhenglian; Zhang, Chong#; Xing, Xin-Hui. Genome-wide screening identifies promiscuous phosphatases impairing terpenoid biosynthesis in Escherichia coli. Applied Microbiology and Biotechnology. November 2018. 102(22):9771-9780.
- 9. Guo, Jiahui*; Wang, Tianmin*; Guan, Changge; Liu, Bing; Luo, Cheng; Xie, Zhen; Zhang, Chong#; Xing Xin-Hui. Improved sgRNA design in bacteria via genome-wide activity profiling. Nucleic Acids Research. August 2018. 46(14):7052-7069.
- 10. Wu, Yinan*; Wang, Tianmin*; Zhang, Chong#; Xing, Xin-Hui. A rapid and specific colorimetric method for free tryptophan quantification. Talanta. January 2018. 176:604-609.
- 11. Fang, Mingyue*; Wang, Tianmin*; Zhang, Chong#; Bai, Jili; Zheng, Xiang; Zhao, Xuejin; Lou, Chunbo; Xing, Xin-Hui. Intermediate-sensor assisted push-pull strategy and its application in heterologous deoxyviolacein production in Escherichia coli. Metabolic Engineering. January 2016. 33:41-51.
- 12. Wang, Tianmin*; Mori, Hiroshi; Zhang, Chong; Kurokawa Ken; Xing Xin-Hui#; Yamada, Takuji#. DomSign: a top-down annotation pipeline to enlarge enzyme space in the protein universe. BMC Bioinformatics. March 2015. 16:96.
- 13. Mo, Xuhua*; Zhang, Hui*; Wang, Tianmin; Zhang, Chong; Zhang, Cong; Xing Xin-Hui; Yang, Song#. Establishment of CRISPR interference in Methylorubrum extorquens and application of rapidly mining a new phytoene desaturase involved in carotenoid biosynthesis. Applied Microbiology and Biotechnology. May 2020. 104(10):4515-4532.
- 14. Liu, Shude*; Wu, Yinan*; Wang, Tianmin; Zhang, Chong#; Xing, Xin-Hui. Maltose Utilization as a Novel Selection Strategy for Continuous Evolution of Microbes with Enhanced Metabolite Production. ACS Synthetic Biology. December 2017. 6(12):2326-2338.
- 15. Liang, Weifan*; Cui, Lanyu; Cui, Jinyu; Yu, Kaiwen; Yang, Song; Wang, Tianmin; Guan Changge; Zhang, Chong#; Xing, Xin-Hui. Biosensor-assisted transcriptional regulator engineering for Methylobacterium extorquens AM1 to improve mevalonate synthesis by increasing the acetyl-CoA supply. Metabolic Engineering. January 2017. 39:159-168.
- 16. Zhang, Shouchi*; Lai, Qiheng; Lu, Yuan; Liu, Zhidan; Wang, Tianmin; Zhang, Chong#; Xing, Xin-Hui. Enhanced biohydrogen production from corn stover by the combination of Clostridium cellulolyticum and hydrogen fermentation bacteria. Journal of Bioscience and Bioengineering. October 2016. 122(4):482-7.
- 17. Feng, Quan*; Wang, Yuxiao; Wang, Tianmin; Zheng, Hao; Chu, Libing; Zhang, Chong; Chen, Hongzhang; Kong, Xiuqin; Xing, Xin-Hui#. Effects of packing rates of cubic-shaped polyurethane foam carriers on the microbial community and the removal of organics and nitrogen in moving bed biofilm reactors. Bioresource Technology. August 2012. 117:201-7.
Awards
- 1. 2018/11, Distinguished Postdoctoral Fellowship, Tsinghua-Peking Center for Life Sciences
- 2. 2019/04, National Postdoctoral Innovation Talent Support Program
|





