Principal investigator
Name:
Bei YangAssistant Professor , PhD, Assistant Professor
Position:
Principle Investigator, Tenure-track Assistant Professor
Affiliation:
Honor:
Oriental Talents Program
Education Background:
- 2000/09-2004/06, National Talents training program, College of Life Science, Wuhan University, B.S.
- 2004/09-2010/07, Shanghai Institute of Biochemistry and Cell Biology (SIBCB), Chinese Academy of Sciences (CAS), Ph.D.
Working Experience:
- 2010/08-2013/06, National Institutes of Health (NIH), Postdoctoral Fellow
- 2013/07-2015/07, University of California at Berkeley, Postdoctoral Fellow
- 2015/11-2020/09, Shanghai Institute for Advanced Immunochemical Studies, ShanghaiTech University, Research Associate Professor
- 2020/10-Present, Shanghai Institute for Advanced Immunochemical Studies, ShanghaiTech University, PI
- 2020/10-Present, School of Life Science and Technology, ShanghaiTech University, Tenure-track Assisitant Professor
Group Introduction
Research Area:
infectious diseases, rare hereditary diseases
Research Interests:
We use an array of different methods, including cryo-EM, mass spectrometry, high-throughput screening, animal models etc., to investigate biological questions with clinical significance. We study the molecular basis of a range of high-risk infectious diseases (e.g. AIDS) and genetic diseases (e.g. β-thalassemia), as well as gene editing tools, so as to identify novel therapeutic and preventive opportunities. On top of the basic research, we also seek to translate gained knowledge into the development of preventive and therapeutic interventions including gene therapies and mRNA vaccines. 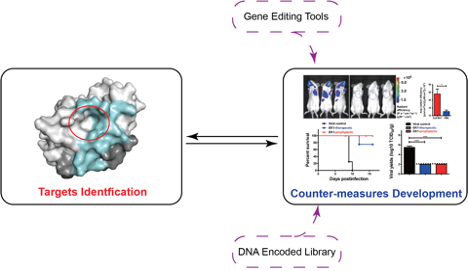
Research Achievement
Our representative research achievements are briefed below: 1. Development and application of gene editing tools: In recent years, nuclear and mitochondrial base editors have been developed by coupling the “locators” like CRISPR/Cas9 or TALE proteins with the “effectors” like APOBEC or DddA to enable base substitutions at defined sites in nuclear and mitochondrial genomes. However, the first-generation of such editors exhibited certain levels of off-target and bystander editing, limiting their broad application in both research and clinical settings. In the past, we have leveraged our expertise in protein engineering to contribute in the development of several high-precision nuclear genome editing tools, including a transformer base editor (tBE) with no detectable off-target effect (Nat Cell Biol, 2021). More recently, we captured the structural snapshots of DdCBE, the mitochondrial base editor, targeting two native mitochondrial gene loci, thus revealing the structural determinants of its editing window. Based on the mechanistic insights, we then developed a “WinPred” model to guide TALE-recognition region and spacer length design and engineered a high-precision variant of DdCBE, i.e., aDdCBE with a 2~3 nt editing window and minimal off-target mutations. Further application of the WinPred model and aDdCBE enabled near single-nucleotide editing at multiple target sites and allowed faithful modelling of Leber hereditary optic neuropathy disease-related mutations (Mol Cell, 2025) (Fig.1). Besides gene editing tools development, we are also actively exploring their application in high-risk infectious disease prevention and hereditary diseases treatment. For instance, we and our collaborators used tBE to disrupt transcription factor binding motifs in the promoter region of γ-globin in hematopoietic stem cells, thereby reactivating the silenced γ-globin expression for β-thalassemia treatment (Cell Stem Cell, 2023).
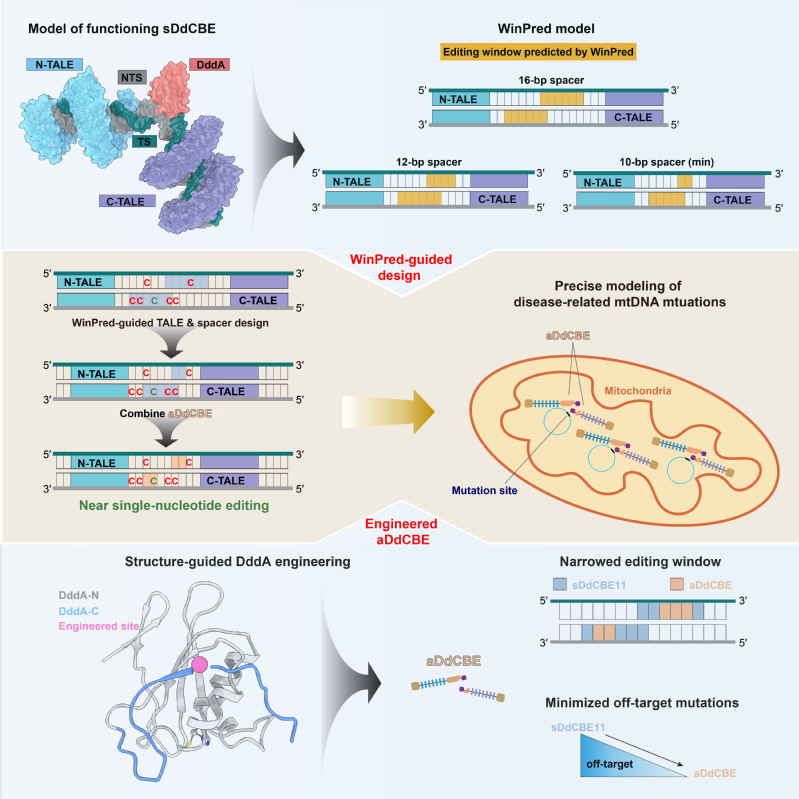
Fig.1| Structure-guided development of high-precision mitochondrial base editors 2. Vaccine targets Identification: Revealing conserved immune vulnerabilities in potential immunogens is essential for effective and broad-spectrum vaccine development. The trimeric Env protein on HIV-1 particles has long been the focus of AIDS vaccine design. However, its substantial sequence variability across HIV-1 subtypes and extensive glycosylation together impeded vaccine development for decades. To enable the application of state-of-the-art “epitope-focusing” vaccine strategy on HIV-1, we characterized the structural and immunogenic landscape of Env proteins from Asia prevalent HIV-1 subtypes, CRF01_AE and CRF07_BC. Our work identified CRF01_AE-specific features in the Env V1 region (Fig. 2A–B), uncovered their association with resistance to certain broadly neutralizing antibodies (bNAbs) (Fig. 2C–D), and revealed a novel neutralization mechanism of the first bNAb isolated from a CRF01_AE-infected individual (Fig. 2E) (Nat Commun, 2023). These findings expanded our understanding of Asia-prevalent HIV-1 subtypes and shed lights on future “epitope-focusing” HIV-1 vaccine design. Meanwhile, to better prepare for potential emergent coronaviruses, i.e., “HCoV-X”, we mapped the antigenic landscape of α-coronavirus spike proteins and revealed both conserved and divergent antigenic features across human coronaviruses, thus providing clues for future development of broadly effective coronavirus vaccines (Commun Biol, 2022).
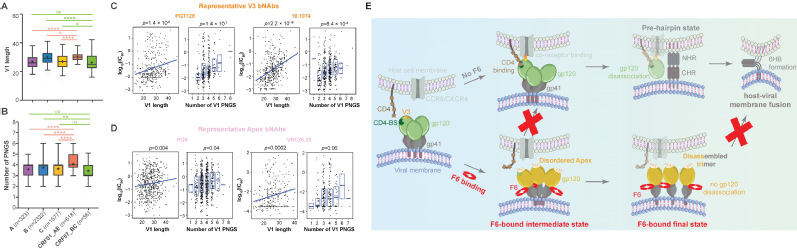
Fig.2| Structural and immunogenic understanding of Envs from CRF subtypes 3. Drug targets identification and inhibitors screening: Compared to the huge health and economic burden imposed by Enterovirus infections on society, the countermeasures have fallen short. Viral 3C protease is one of the drug development focuses of Enterovirus, yet its protease active center has frustrated previous drug discovery efforts. Using an integrative approach which combines mass spectrometry and X-ray crystallography, we demonstrated that 1) the protease activity of 3C could be modulated allosterically and 2) 3C plays a regulatory role in enteroviruses genome replication by binding to the 5’NCR of viral genome. Notably, both the allosteric site and the 5'NCR-binding site on 3C are highly conserved and locate away from the protease active center, making them attractive broad-spectrum drug targets for enterovirus infection control (PNAS, 2020). In silico screening of 143,621 natural products against the allosteric site further identified dihydromyricetin (DHM), a natural product that can broadly binds and allosterically inhibits the protease activities of 3C proteins from multiple enteroviruses. Notably, DHM shows minimal cytotoxicity and potent antiviral efficacy in different cell models, exhibiting a selective index exceeding 700. These findings establish DHM as a unique, broad-spectrum allosteric inhibitor of Enterovirus 3C proteases and underscore its potential as a promising candidate for the development of pan-enterovirus antivirals (Adv Biol, 2025).
Representative Publications (*First Author, # Corresponding Author)
- 1. Jiangchao Xiang*, Wenchao Xu*, Jing Wu*, Yaxin Luo, Chengyu Liu, Yaofeng Hou, Jia Chen, Bei Yang.Structural insights into DdCBE in action enable highprecision mitochondrial DNA editing.MOLECULAR CELL. 10 Sep 2025.
- 2. Gao, Yan*; Xie, Xiong; Zhang, Xiaoyu; Cao, Junyuan; Lan, Weiqi; You, Tian; Li, Dongxu; Dong, Xuxue; Dai, Wenhao; Xiang, Yingchun; Hu, Shulei; Shang, Weijuan; Wu, Botao; Zhang, Yumin; Xu, Jin; Liu, Xiaoce; Wang, Haofeng; Hu, Wanlong; Zhang, Mingjing; Duan, Yinkai; Cui, Wen; Zhou, Hao; Mao, Shengjiang; Jia, Handi; Sun, Zhanqi; Jia, Menghan; Yin, Yue; Nguyen, Henry C.; Yang, Kailin; Yang, Bei; Yang, Xiuna; Ji, Xiaoyun; Xiao, Gengfu; Wang, Wei; Zhang, Leike; Rao, Zihe; Liu, Hong; Yang, Haitao.Substrate recognition and cleavage mechanism of the monkeypox virus core protease.NATURE. 01 Apr 2025.
- 3. Chen, Jiayan*; Wang, Qi; He, Xiaomeng; Yang, Bei.Malaria Vaccines: Current Achievements and Path Forward.VACCINES. 19 May 2025. 13(5).
- 4. Sun, Shangwu*; Wang, Qiang; Zhu, Mengyao; Zhang, Xuan; Zhang, Xianfang; Yang, Bei.A Pan-Enterovirus Natural Product Inhibitor Targeting a Unique Allosteric Site on the Viral 3C Protease.ADVANCED BIOLOGY. 01 Jul 2025.
- 5. Guangye Li*; Guo Chen*; Guo-Hua Yuan*; Jia Wei*; Qingyang Ni; Jing Wu; Bei Yang; Li Yang; Jia Chen.Specific and efficient RNA A-to-I editing through cleavage of an ADAR inhibitor.NATURE BIOTECHNOLOGY. 26 Mar 2025.
- 6. Fan, Yuhang*; Xu, Wenchao; Gao, Bao-Qing; Qin, Huichao; Wu, Xiaoyi; Wei, Jia; Ni, Qingyang; Zhou, Lina; Xiang, Jiangchao; Wu, Jing; Yang, Bei; Yang, Li; Chen, Jia.Leveraging base excision repair for efficient adenine base editing of mitochondrial DNA.NATURE BIOTECHNOLOGY. 25 Mar 2025.
- 7. Zhang, Ruiwen*; He, Zhou; Shi, Yajing; Sun, Xiangkun; Chen, Xinyu; Wang, Guoquan; Zhang, Yizhou; Gao, Pan; Wu, Ying; Lu, Shuhan; Duan, Junyi; Sun, Shangwu; Yang, Na; Fan, Wei; Zhao, Kaitao; Yang, Bei; Xia, Yuchen; Zhang, Yan; Zhang, Ying; Yin, Hao.Amplification editing enables efficient and precise duplication of DNA from short sequence to megabase and chromosomal scale.CELL. 25 Jul 2024. 187(15).
- 8. Luo, Yaxin*; Hou, Yaofeng; Zhao, Wenwen; Yang, Bei.Recent progress in gene therapy for familial hypercholesterolemia treatment.ISCIENCE. 20 Sep 2024. 27(9).
- 9. Han, Wenyan*; Qiu, Hou-Yuan*; Sun, Shangwu*; Fu, Zhi-Can*; Wang, Guo-Quan*; Qian, Xiaowen*; Wang, Lijie; Zhai, Xiaowen; Wei, Jia; Wang, Yichuan; Guo, Yi-Lin; Cao, Guo-Hua; Ji, Rui-Jin; Zhang, Yi-Zhou; Ma, Hongxia; Wang, Hongsheng; Zhao, Mingli; Wu, Jing; Bi, Lili; Chen, Qiu-Bing; Li, Zifeng; Yu, Ling; Mou, Xiaodun; Yin, Hao; Yang, Li; Chen, Jia; Yang, Bei; Zhang, Ying.Base editing of the HBG promoter induces potent fetal hemoglobin expression with no detectable off-target mutations in human HSCs.CELL STEM CELL. Nov 2023.
- 10. Niu, Jun*; Wang, Qi; Zhao, Wenwen; Meng, Bing; Xu, Youwei; Zhang, Xianfang; Feng, Yi; Qi, Qilian; Hao, Yanling; Zhang, Xuan; Liu, Ying; Xiang, Jiangchao; Shao, Yiming; Yang, Bei.Structures and immune recognition of Env trimers from two Asia prevalent HIV-1 CRFs.NATURE COMMUNICATIONS. 04 Aug 2023. 14(1).
- 11. Zhao, Wenwen*; Li, Jifang*; Wang, Xiao*; Xu, Wei*; Gao, Bao-Qing*; Xiang, Jiangchao; Hou, Yaofeng; Liu, Wei; Wu, Jing; Qi, Qilian; Wei, Jia; Yang, Xiaoyu; Lu, Lu; Yang, Li; Chen, Jia; Yang, Bei.Prime editor-mediated functional reshaping of ACE2 prevents the entry of multiple human coronaviruses, including SARS-CoV-2 variants.MEDCOMM. 2023.
- 12. Shangwu Sun*; Rui Zhu*; Mengyao Zhu; Qi Wang; Na Li; Bei Yang.Visualization of conformational transition of GRP94 in solution.LIFE SCIENCE ALLIANCE. Nov 2023. 7(2).
- 13. Jiangchao Xiang*; Wenchao Xu*; Jing Wu*; Yaxin Luo; Bei Yang; Jia Chen.Nucleoside deaminases: the key players in base editing toolkit.BIOPHYSICS REPORTS. 01 Nov 2023.
- 14. Jia, Xinshuo*; Li, Yanan*; Wang, Teng; Bi, Lulu; Guo, Lijuan; Chen, Ziting; Zhang, Xia; Ye, Shasha; Chen, Jia; Yang, Bei; Sun, Bo.Discrete RNA–DNA hybrid cleavage by the EXD2 exonuclease pinpoints two rate-limiting steps.THE EMBO JOURNAL. 04 Jan 2023. 42:e111703.
- 15. Xiang, Jiangchao*; Su, Jie; Lan, Qiaoshuai; Zhao, Wenwen; Zhou, Yu; Xu, Youwei; Niu, Jun; Xia, Shuai; Qi, Qilian; Sidhu, Sachdev; Lu, Lu; Miersch, Shane; Yang, Bei.Antigenic mapping reveals sites of vulnerability on alpha-HCoV spike protein.COMMUNICATIONS BIOLOGY. 04 Nov 2022. 5(1).
- 16. Li, Xiaosa*; Zhou, Lina*; Gao, Bao-Qing*; Li, Guangye; Wang, Xiao; Wang, Ying; Wei, Jia; Han, Wenyan; Wang, Zixian; Li, Jifang; Gao, Runze; Zhu, Junjie; Xu, Wenchao; Wu, Jing; Yang, Bei; Sun, Xiaodong; Yang, Li; Chen, Jia.Highly efficient prime editing by introducing same-sense mutations in pegRNA or stabilizing its structure.NATURE COMMUNICATIONS. 29 Mar 2022. 13(1):1669.
- 17. Wang, Lijie*; Xue, Wei*; Zhang, Hongxia*; Gao, Runze*; Qiu, Houyuan*; Wei, Jia; Zhou, Lina; Lei, Yun-Ni; Wu, Xiaocheng; Li, Xiao; Liu, Chengfang; Wu, Jing; Chen, Qiubing; Ma, Hanhui; Huang, Xingxu; Cai, Cheguo; Zhang, Ying; Yang, Bei; Yin, Hao; Yang, Li; Chen, Jia.Eliminating base-editor-induced genome-wide and transcriptome-wide off-target mutations.NATURE CELL BIOLOGY. May 2021. 23(5):552-563.
- 18. Xiaojie Shi*; Yue Wan*; Nan Wang*; Jiangchao Xiang*; Tao Wang; Xiaofeng Yang; Ju Wang; Xuxue Dong; Liang Dong; Lei Yan; Yu Li; Lili Liu; Shinchen Hou; Zhenwei Zhong; Ian A. Wilson; Bei Yang; Guang Yang; Richard A. Lerner.Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms.NATURE COMMUNICATIONS. 05 May 2021.
- 19. Meng, Bing*; Lan, Keke; Xie, Jia; Lerner, Richard A.; Wilson, Ian A.; Yang, Bei.Inhibitory antibodies identify unique sites of therapeutic vulnerability in rhinovirus and other enteroviruses.PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA. 16 Jun 2020. 117(24):13499-13508.
- 20. Wang, Xiao*; Ding, Chengfeng*; Yu, Wenxia*; Wang, Ying*; He, Siting*; Yang, Bei*; Xiong, Yi-Chun; Wei, Jia; Li, Jifang; Liang, Jiayi; Lu, Zongyang; Zhu, Wei; Wu, Jing; Zhou, Zhi; Huang, Xingxu; Liu, Zhen; Yang, Li; Chen, Jia.Cas12a Base Editors Induce Efficient and Specific Editing with Low DNA Damage Response.CELL REPORTS. 02 Jun 2020. 31(9).
- 21. Cui, Yan-ru*; Wang, Shao-jie*; Chen, Jun; Li, Jie; Chen, Wenzhang; Wang, Shuyue; Meng, Bing; Zhu, Wei; Zhang, Zhuhong; Yang, Bei; Jiang, Biao; Yang, Guang; Ma, Peixiang; Liu, Jia.Allosteric inhibition of CRISPR-Cas9 by bacteriophage-derived peptides.GENOME BIOLOGY. 26 Feb 2020. 21(1).
- 22. Xu, Juncao*; Cui, Kaijie*; Shen, Liqiang; Shi, Jing; Li, Lingting; You, Linlin; Fang, Chengli; Zhao, Guoping; Feng, Yu; Yang, Bei; Zhang, Yu.Crl activates transcription by stabilizing active conformation of the master stress transcription initiation factor.ELIFE. 17 Dec 2019. 8.
- 23. Yang, Li*; Yang, Bei; Chen, Jia.One Prime for All Editing.CELL. Dec 2019. 179(7):1448-1450.
- 24. Zhao, Quanju*; Ren, Chaowei*; Liu, Linyi*; Chen, Jinju; Shao, Yubao; Sun, Ning; Sun, Renhong; Kong, Ying; Ding, Xinyu; Zhang, Xianfang; Xu, Youwei; Yang, Bei; Yin, Qianqian; Yang, Xiaobao; Jiang, Biao.Discovery of SIAIS178 as an Effective BCR-ABL Degrader by Recruiting Von Hippel-Lindau (VHL) E3 Ubiquitin Ligase.JOURNAL OF MEDICINAL CHEMISTRY. 24 Oct 2019. 62(20):9281-9298.
- 25. Wang, Ying*; Gao, Runze*; Wu, Jing*; Xiong, Yi-Chun; Wei, Jia; Zhang, Sipin; Yang, Bei; Chen, Jia; Yang, Li.Comparison of cytosine base editors and development of the BEable-GPS database for targeting pathogenic SNVs.GENOME BIOLOGY. Oct 2019. 20(1).
- 26. Chen, Jia*; Yang, Bei; Yang, Li.To BE or not to BE, that is the question.NATURE BIOTECHNOLOGY. May 2019. 37(5):520-521.
- 27. Xia, Shuai*; Yan, Lei*; Xu, Wei*; Agrawal, Anurodh Shankar; Algaissi, Abdullah; Tseng, Chien-Te K.; Wang, Qian; Du, Lanying; Tan, Wenjie; Wilson, Ian A.; Jiang, Shibo; Yang, Bei; Lu, Lu.A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike.SCIENCE ADVANCES. Apr 2019. 5(4).
- 28. Yang, Bei*; Yang, Li; Chen, Jia.Development and Application of Base Editors.CRISPR JOURNAL. Apr 2019. 2(2):91-104.
- 29. Wang, Xiao*; Li, Jianan*; Wang, Ying*; Yang, Bei*; Wei, Jia*; Wu, Jing; Wang, Ruixuan; Huang, Xingxu; Chen, Jia; Yang, Li.Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion.NATURE BIOTECHNOLOGY. Oct 2018. 36(10):946-949.
- 30. Yan, Lei*; Meng, Bing; Xiang, Jiangchao; Wilson, Ian A.; Yang, Bei.Crystal structure of the post-fusion core of the Human coronavirus 229E spike protein at 1.86 angstrom resolution.ACTA CRYSTALLOGRAPHICA SECTION D-STRUCTURAL BIOLOGY. Sep 2018. 74:841-851.
- 31. Qiang, Min*; Dong, Xue*; Zha, Zhao; Zuo, Xiao-Kun; Song, Xing-Lei; Zhao, Lixia; Yuan, Chao; Huang, Chen; Tao, Pingdong; Hu, Qin; Li, Wei-Guang; Hu, Wanhui; Li, Jie; Nie, Yan; Buratto, Damiano; Zonta, Francesco; Ma, Peixiang; Yu, Zheng; Liu, Lili; Zhang, Yi; Yang, Bei; Xie, Jia; Xu, Tian-Le; Qu, Zhihu; Yang, Guang; Lerner, Richard A..Selection of an ASIC1a-blocking combinatorial antibody that protects cells from ischemic death.PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA. 07 Aug 2018. 115(32):E7469-E7477.
- 32. Li, Xiaosa*; Wang, Ying*; Liu, Yajing*; Yang, Bei*; Wang, Xiao; Wei, Jia; Lu, Zongyang; Zhang, Yuxi; Wu, Jing; Huang, Xingxu; Yang, Li; Chen, Jia.Base editing with a Cpf1-cytidine deaminase fusion.NATURE BIOTECHNOLOGY. Apr 2018. 36(4):324-327.
- 33. Lei, Liqun*; Chen, Hongquan*; Xue, Wei*; Yang, Bei*; Hu, Bian*; Wei, Jia; Wang, Lijie; Cui, Yiqiang; Li, Wei; Wang, Jianying; Yan, Lei; Shang, Wanjing; Gao, Jimin; Sha, Jiahao; Zhuang, Min; Huang, Xingxu; Shen, Bin; Yang, Li; Chen, Jia.APOBEC3 induces mutations during repair of CRISPR-Cas9-generated DNA breaks.NATURE STRUCTURAL & MOLECULAR BIOLOGY. Jan 2018. 25(1):45-52.
- 34. Wang, Lijie*; Xue, Wei*; Yan, Lei*; Li, Xiaosa; Wei, Jia; Chen, Miaomiao; Wu, Jing; Yang, Bei; Yang, Li; Chen, Jia.Enhanced base editing by co-expression of free uracil DNA glycosylase inhibitor.CELL RESEARCH. Oct 2017. 27(10):1289-1292.
- 35. Yang, Bei*; Li, Xiaosa; Lei, Liqun; Chen, Jia.APOBEC: From mutator to editor.JOURNAL OF GENETICS AND GENOMICS. Sep 2017. 44(9):423-437.
Funding
- 1. 2024-General Program, Natural Science Foundation of China, Leader
- 2. 2023-General Program, Science and Technology Commission of Shanghai Municipality, Leader
- 3. 2023-Major Program , Ministry of Science and Technology of China, Participator
- 4. 2021-General Program, Natural Science Foundation of China, Leader
- 5. 2019-Key Program , Ministry of Science and Technology of China, Participator
- 6. 2018-Major Program , Ministry of Science and Technology of China, Participator
- 7. Youth Program, Natural Science Foundation of China, Leader
Awards
- 1. 2023, Oriental Talents Program
- 2. 2016, Pujiang Talents Program
Group Member and Photo
-
Name:Qilian Qi
Position:Engineer
Duration:2016/01~present
Email:qiql@@shanghaitech.edu.cn
-
Name:Xianfang Zhang
Position:Engineer
Duration:2016/07~present
Email:zhangxf1@@shanghaitech.edu.cn
-
Name:Qi Wang
Position:Doctoral Student
Duration:2020 ~ present
Email:wangqi3@@shanghaitech.edu.cn
-
Name:Yanxin Luo
Position:Doctoral Student
Duration:2021 ~ present
Email:luoyx1@@shanghaitech.edu.cn
-
Name:Yaofeng Hou
Position:Doctoral Student
Duration:2022 ~ Present
Email:houyf2022@@shanghaitech.edu.cn
-
Name:Qiang Wang
Position:Doctoral Student
Duration:2023 ~ present
Email:wangqiang2023@@shanghaitech.edu.cn
-
Name:Jiayan Chen
Position:Doctoral Student
Duration:2023 ~ present
Email:chenjy22023@@shanghaitech.edu.cn
-
Name:Xiaomeng He
Position:Postgraduate Student
Duration:2024-Now
Email:hexm2024@@shanghaitech.edu.cn
-
Name:Xiaotian Chen
Position:Postgraduate Student
Duration:2024-Now
Email:chenxt2024@@shanghaitech.edu.cn
-
Name:Xindi Shang
Position:Postgraduate Student
Duration:2024-Now
Email:shangxd2024@@shanghaitech.edu.cn
|
















