A team of scientists led by Dr. Jia Chen and Dr. Xingxu Huang of the School of Life Science and Technology at ShanghaiTech University, along with Dr Li Yang of the CAS-MPG Partner Institute for Computational Biology at the Chinese Academy of Sciences, have developed a series of novel CRISPR/Cpf1 (Cas12a) bases editors. The paper entitled “Base editing with a Cpf1– cytidine deaminase fusion” was published online in Nature Biotechnology on March 19, 2018.
The base editors (BEs) developed by combining apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like (APOBEC) or activation-induced deaminase (AID) cytidine deaminase family memberswith the CRISPR-Cas system has been used for targeted C-to-T base editing in various species. These previously developed BEs are dependent on Cas9 proteins for genome targeting, which limits base editing to regions with G/C rich protospaceradjacent motif (PAM) sequences.
In this study, researchers developed a novel BE system based on catalytically dead Lachnospiraceae bacterium Cpf1 (Cas12a).Cpf1 is another Cas protein that differs from Cas9 in several ways: Cpf1 requires a T-rich PAM sequence (TTTV) for target- DNA recognition; the guide RNA for Cpf1 (CRISPR RNA (crRNA)) is shorter than that for Cas9 (single guide RNA (sgRNA)); and the Cpf1-cleavage site is in a distal and downstream location relative to the PAM sequence in spacer DNA, rather than a proximal and upstream one as in Cas9. Cpf1 also induces less off-target cleavage genome-wide than does Cas9. These newly developed dCpf1-BEs perform targeted base editing with very low levels of indel formation, non-C-to-T substitutions and off-target editing, thus enabling base editing in A/T-rich regions with high precision.
This study was supported by grants from the National Natural Science Foundation of China, Ministry of Science and Technology, Shanghai Municipal Science and Technology Commission, and ShanghaiTech University.
Read more at:https://www.nature.com/articles/nbt.4102
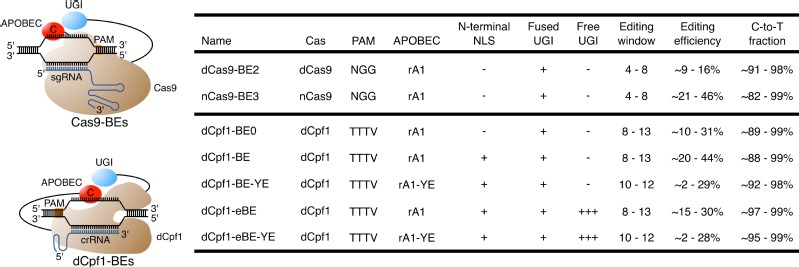
Summary of Cas9-based and dCpf1-based BEs


