Cell and cellular organelles communication are often controlled by the membrane protein interaction. Despite the many existing methods to detect protein-protein interactions (PPIs), there are still challenges in detecting membrane PPIs. Transient and weak PPIs are mostly associated with membrane receptor-mediated signaling pathways. Mass spectrometry-based membrane protein interaction studies have also not been widely successful, due to the poor solubility of membrane proteins or the loss of PPIs during sample preparation. In recent years, protein proximity-tagging methods, such as APEX and BioID, have been applied in membrane protein studies with great results.
School of Life Science and Technology Professor Min Zhuang’s group has developed a novel method whichspecifically tags the interacting “membrane proteins of interest” (MPOI) in cells. Their research regarding this new method, called PUP-IT (Pupylation-based interaction tagging, was published online on August 13th, 2018, in Nature Methods. This research offers an alternative proximity-tagging system to study protein-protein interactions. This approach transforms transient and weak interactions into others that involve covalent binding, so that the tagged proteins can be subsequently enriched by affinity purification under denaturing conditions for mass spectrometry-based identification.
In the paper, the researchers first applied this approach to CD28, a critical co-stimulatory receptor for T lymphocyte activation. Three known CD28 binding partners and multiple potential interacting proteins were identified. In addition, this system was proven efficient in tracking cell-cell interactions. Finally, they showed that this method can be used to identify the interaction between a cell surface receptor and its ligand. The paper demonstrated a powerful tool uncovering the weak and transient membrane PPIs in cells, which will have a broad application in the future.
Graduate students Qiang Liu and Jun Zheng are first co-authors. Professors Min Zhuang and Haopeng Wang are correspondingco-authors. Their work has been mainly supported by ShanghaiTech and the National Natural Science Foundation of China.
Read more at: https://www.nature.com/articles/s41592-018-0100-5
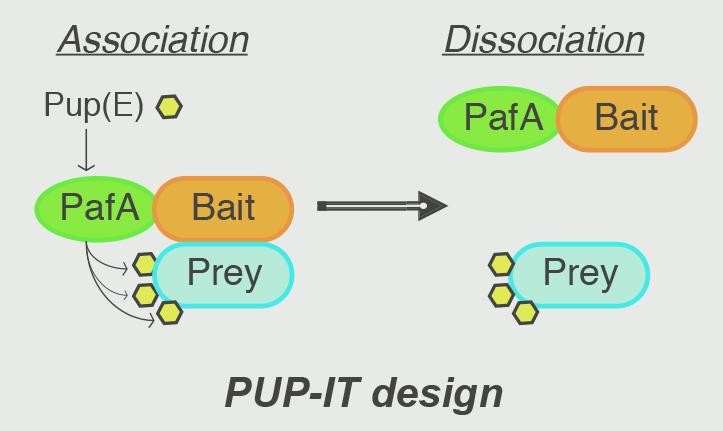
Sketch map of PUT-IT work principle


