In a new paper published in Developmental Cell, a team of researchers from ShanghaiTech University, Shenzhen Institutes of Advanced Technology, Tsinghua University, Johns Hopkins University and National Institute of Biological Sciences developed a powerful strategy and valuable reagents to systematically probe histone functions in Drosophila melanogaster.
Associate Professor Gao Guanjun from the School of Life Science and Technology at ShanghaiTech University and Professor Dai Junbiao from Shenzhen Institutes of Advanced Technology are the senior authors of the study, entitled “Probing the function of metazoan histones with a systematic library of H3 and H4 mutants,” which appeared online in the journal on December 28th, 2018. Professor Gao is the Lead Contact of the paper. The paper’s lead authors are Zhang Weimin, Zhang Xuedi, Xue Zhaoyu and Li Yijie.
Histone post-translational modifications (PTMs), including acetylation, methylation, phosphorylation, ubiquitination, and crotonylation, are thought to modulate chromatin structure and gene expression either directly or via recruitment of specific chromatin-associated proteins. Studies involving genetic or chemical interventions targeting histone-modifying enzymes have provided substantial evidence for biological functions of specific PTMs. However, investigating histone functions has been extremely challenging for the following reasons. First, these modifying enzymes generally have protein substrates other than histones. Second, chromatin-regulating enzymes have additional functions unrelated to enzymatic activities. Third, the roles of PTMs can be directly queried by systematic mutation of histone residues, which in higher organisms pose additional challenges. Four, complexity of multiple copies and multiple distributions of histone genes in multicellular organisms makes it difficult to substantially alter levels of particular histone proteins substantially.
In this study, researchers generated an efficient histone-mutagenesis platform, enabling the functional study of each residue in all five histones with much higher throughput than previous techniques. As a proof-of-concept study, researchers targeted H3 and H4, systematically analyzing the functional relationships between histone modification sites and growth, development, DNA damage, transposon activity, maintenance and differentiation of germline stem cells.
This work was supported by National Natural Science Foundation of China, National Key Research and Development Program of China, NIH-R01 and start-up funding from ShanghaiTech University.
Read more at: https://www.cell.com/developmental-cell/fulltext/S1534-5807(18)31031-1
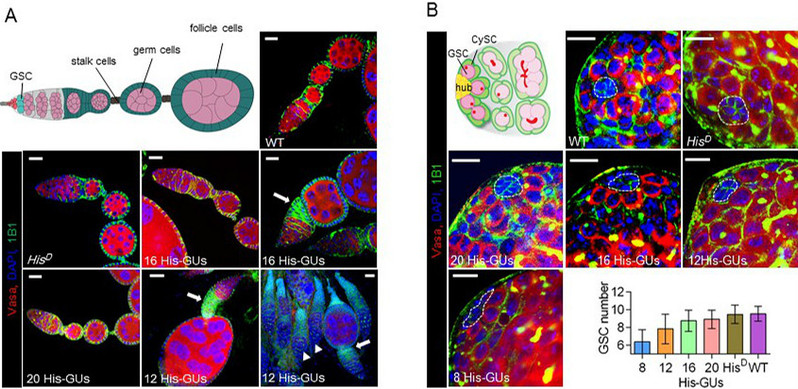
Low histone dosage affects testis and ovary development


